|
| List of Stored Contents | |||||
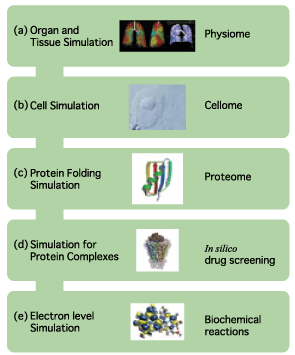
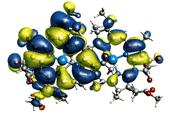
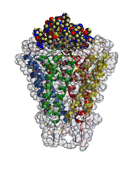
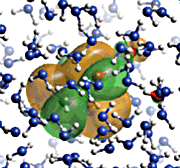
| ♦Electron level simulation 1 AMOSS (Ab initio Molecular Orbital Simulation on Supercomputer) The quantum chemical program for computing ab initio molecular orbitals developed by the NEC Fundamental Research Laboratories offers fast, high-accuracy computation under the condition of a vast array of systems. The CI (Configuration Interaction) computation method is also under development. (figure 1) ♦Electron level simulation 2 DFT (Density Function Theory)/Generalized DFT The program for DFT and generalized DFT, developed by the group led by Prof. Kizashi YAMAGUCHI, Graduate school of Science, Osaka Univ., can compute the electron states of complicated systems involving spin frustration, spin degeneracy, and spin canting. Thus, simulations are available for biochemical reactions, in particular, of complicated reactions involving oxidization and reduction. ♦Simulation for Protein Complexes prestoX-basic The molecular dynamics program is being developed in cooperation with JBIRC, Institute for Protein Research, Osaka Univ., Yokohama City University, Hitachi Corp. and Fujitsu Corp. for achievement of in silico drug screening. (figure 2) ♦Protein Fold Simulation Prediction of ab initio protein folds from amino acid sequence information The group led by Prof. Shoji TAKADA, Faculty of Science, Kobe Univ., develops a sophisticated algorithm to predict protein folds, and their program has received high commendation from the international prediction contest "CASP." ♦Cell Simulation Simulation to study the drug inhibitions to ion channels in human cardiac muscle cells. The study is being conducted by Prof. Yoshihisa KURACHI, Graduate School of Medicine, Osaka Univ. ♦Organ and Tissue Simulation Simulations of surface muscle electric potential and lung ventilation These studies at the organ and tissue levels are performed by Prof. Kenzo AKAZAWA and Dr. Hiroko KITAOKA, Graduate School of Information Science and Technology. Osaka Univ. They will offer an appropriate diagnosis and medical treatment. |
|
||||
| A new platform for performing coupled computations by integrating application programs at different levels on the grid, which consists of various types of heterogeneous computers. In particular, the platform focusing on biosimulation is referred to as BioPfuga. | |||||||||
|
|
||||||||
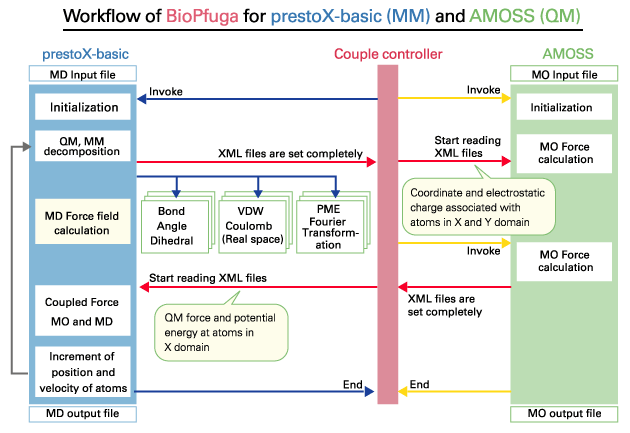 |
|||||||||
|
| The biological simulation programs will be applied to these developments at the levels of electrons, biomolecules and cells/organs to demonstrate their quality following the paradigm of life science. |
| Top | Project | Research work | Archive | Event | Link | |