 |
 |
| As represented by the term "Bioinformatics," advances in information
science and technology are expected to be applied to the field of life science.
There are a vast number of biological databases containing biological information,
in particular, more than 500 databases for genomic information. These databases
have been operated and managed individually on the Internet. Under such a circumstance,
we have founded an infrastructure that allows to share information contained in
these databases and to conduct research collaboration. |
| However, the following problems exist in interoperating these databases. |
- Difference in format depending on database
- Necessity of the latest data against the large size of databases and high
frequency of their update.
|
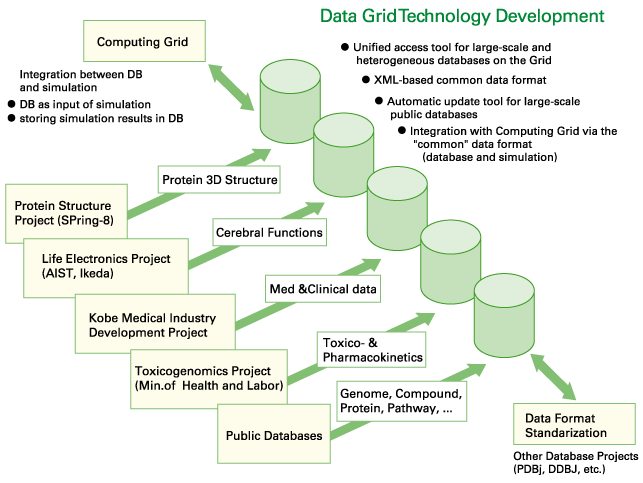
Physical integration of the databases is not practical to solve these problems.
This group is developing several fundamental technologies for making full use
of biological information by the seamless federation of multi-scale databases
with the Grid technology. |
 |
 |
| Seamless Federation of Muli-Scale Databases
with the Grid Technology |
|
 |
| 1. Solving the problem of heterogeneous database
formats XML standard format |
| n the biological databases available on the Web, their database format has
been shifted from the flat file to the XML format. However, these formats have
heterogeneity in their description even for the same protein. For example, figure
shows the RbsB protein, a ribose binding protein, is differently represented in
the DDBJ, SWISS-PROT and PDB databases. |
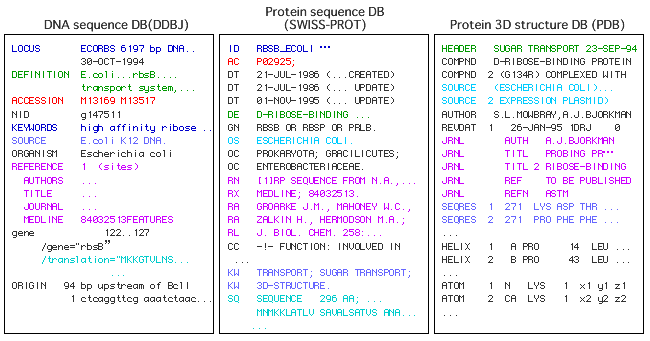 |
| Heterogenity of Bio-Databases(Flat Files) |
|
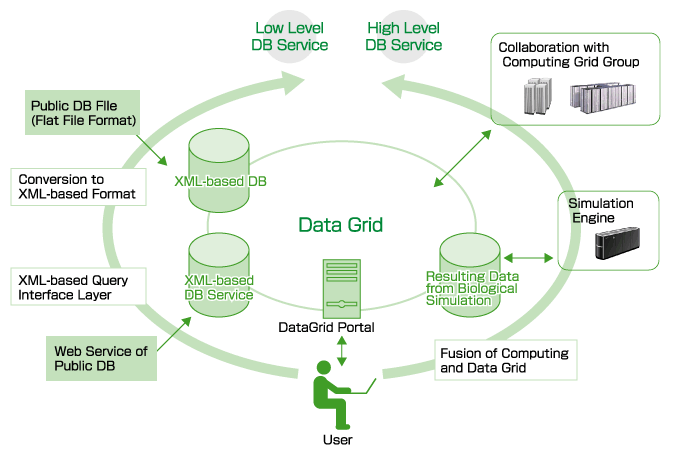
To cope with this issue, a database conversion system has developed for transforming
heterogeneous database formats with different schemes into an XML-based standard
format using conversion rules, and for displaying them.
(Associate Prof. Takenao OHKAWA, Graduate School of Information Science and Technology,
Osaka University; in cooperation with Aztec System Co., Ltd.)
In addition, to achieve seamless search with databases distributed in the network
(making use of the independence of the different database management policies),
the prototype of the Grid Database Infrastructure System is being developed so
that the databases will be integrated virtually using Web mechanisms such as SOAP
to enable database services to provide within the XML scheme. (Hitachi Software
Engineering Co., Ltd.) |
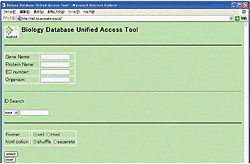
<Unified access tool for biology databases (prototype)>
This tool can retrieve data with keywords (e.g., "HGF" as a gene name)
through a standardized XML format data as the data having the same structure.
The retrieval is not only performed across different database entries having the
keywords, but also, conducted repeatedly against the databases using the protein
sequences in the already-retrieved entries. |
 |
|
| 2. Development of automatic update system for public
databases |
Comprehensive accessibility to the latest data of genomic databases is directly
related to the success of projects. At present, however, 15 million base sequences
are accumulated in the databases everyday on average. To cope with this issue,
a system is being developed for performing an efficient all-inclusive sequence
search by constructing the local copies of public databases and updating them.
(Prof. Teruo YASUNAGA, Genetic Information Research Center, Osaka University;
in cooperation with Hewlett-Packard Japan, Ltd.) |
| 3. Development of a database system with classified
sequences for homology search |
| Problems have arisen with Biodatabase such as enlarged redundancy due not
only to a rapid increase in the data amount, but also the submission of a large
amount of data on specific types of sequences (for example, many alternative sequences
of the HIV protease). This problem is also amplified by the rapid increase of
low quality data due to the massive submission of a large amount of fragment data
such as draft sequences. To solve these problems, this group has developed a new
system using the following two methods. |
- Comparison is made among all the sequence data in a database and a hierarchy
of the results of database search is dynamically built up to minimize any redundancy
in search.
- High-quality database information (secondary information) produced by experts
is added to the output results to allow more accurate information-browsing.
(Prof. Hiroyuki TOH, Bioinformatics Center, The Institute for Chemical Research,
Kyoto University; in cooperation with Mitsubishi Space Software Co., Ltd.)
|
| 4. High-level information retrieval system: XML-based
ADME information database system |
| The system is being built to allow high-level information retrieval on the
relationships with binding affinity between proteins and compounds, associated
with information on drug metabolism by representing it as XML-based ADME information
(Absorption, Distribution, Metabolism and Excretion of drugs), which are important
in the area of new drug design, and linking among the XML-based information on
genes, proteins and compounds. (Fujitsu Kyushu System Engineering Limited) |
| 5. Coordination with existing Web services: Design
of an XML layer |
The system is being developed for interoperation with existing database services
on the Web with minimal customization by intermediating between the services and
the Data Grid with an XML layer. (Protein Research Foundation)
This system may have the following two advantages to the database service providers: |
- The need for copying all the data contained in a database is eliminated.
This means that no update of copies is required.
- Simply by conducting access restriction to the database with several levels,
various types of services can be provided according to the level of customers.
For this reason, the system may be expanded to actual business.
|
 |
| The future perspective of this project is described below. |
- Completion of the DataGrid Infrastructure System
- Interoperation of further heterogeneous databases
- Collaboration with Computing Grid group Results from the simulations conducted
by Computing Grid group will be stored in databases.
|
|



